In 2001, when the first draft of the human genome was completed, researchers were surprised to learn that only 2% of the human genome codes for proteins. At the time, scientists were very focused on proteins and thought that there would be a much larger number of protein-coding genes in the human genome due to our complexity. The term "junk DNA" has been used to describe the other 98% of the genome. With only 20,000 protein-coding genes, the human genome contains almost the same number of genes as the simple roundworm and model organism C. elegans. However, C. elegans has very little excess DNA, suggesting that this junk DNA could be part of the explanation for the increased complexity of humans. This is the starting point for Nessa Carey's second book Junk DNA: A Journey Through The Dark Matter of the Genome, which explains the importance of the non-coding portion of the genome.
In 2001, when the first draft of the human genome was completed, researchers were surprised to learn that only 2% of the human genome codes for proteins. At the time, scientists were very focused on proteins and thought that there would be a much larger number of protein-coding genes in the human genome due to our complexity. The term "junk DNA" has been used to describe the other 98% of the genome. With only 20,000 protein-coding genes, the human genome contains almost the same number of genes as the simple roundworm and model organism C. elegans. However, C. elegans has very little excess DNA, suggesting that this junk DNA could be part of the explanation for the increased complexity of humans. This is the starting point for Nessa Carey's second book Junk DNA: A Journey Through The Dark Matter of the Genome, which explains the importance of the non-coding portion of the genome. Some scientists have argued that the term junk DNA should be scrapped for a more neutral term like non-coding DNA. They suggest that the term is dated and inaccurate. In addition, calling it junk is rather pejorative and is based on the protein-focused view of the genome. Carey's book nicely demonstrates that the other 98% isn't always junk.
Some scientists have argued that the term junk DNA should be scrapped for a more neutral term like non-coding DNA. They suggest that the term is dated and inaccurate. In addition, calling it junk is rather pejorative and is based on the protein-focused view of the genome. Carey's book nicely demonstrates that the other 98% isn't always junk.What is the other 98% of the genome good for then? Some non-coding DNA has well-established functions. For example, the centromeres are the stretches of DNA that allow the chromosomes to attach to the cell's chromosome segregation apparatus (the mitotic spindle) when the cell copies and divides its DNA. Another example is the telomeres, the lengthy repeat regions of DNA at the ends of the chromosomes. Because telomeres shorten with every cell division, they are linked with aging.
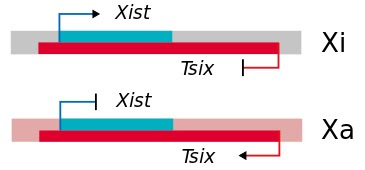
Junk DNA also encodes several special types of RNAs, including long non-coding RNA (lncRNA), microRNA (miRNA), and small interfering RNA (siRNA), that control gene expression. One of the earliest described examples of these special RNAs is found in the biology of sex determination. In XX females, one X chromosome is inactivated to ensure that genes on the X chromosome are not overexpressed. This process, called X chromosome inactivation, is controlled by a gene called Xist (X-inactive specific transcript). Xist encodes a long non-coding RNA, which covers one X chromosome and inactivates it (Xi). Interestingly, on the opposite strand from Xist is a gene called Tsix, which is expressed on the active X chromosome (Xa). The expression of these genes is mutually exclusive, ensuring that only one X chromosome is activated. The Xist/Tsix story highlights the power of special RNAs in controlling gene expression. These RNAs are the subject of intense research in both basic and clinical settings. Carey describes several approved drugs and promising clinical trials based on anti-sense approaches.
In short, Junk DNA was quite readable and should be informative for readers at any level of knowledge about molecular biology. My only complaint about the book was Carey's decision not to include protein or gene names in her writing. In the first chapter, she explains that this was due to the fact that half of her readers find it disruptive. Instead, where applicable, she includes footnotes with the gene or protein names. Unfortunately for me, I am in the half that finds it disruptive to read footnotes to learn the name of the gene in question. Otherwise, the book was very up to date and comprehensive. I also liked her use of simple graphics to explain complex concepts in molecular biology. I recommend Junk DNA for those who want to learn more about why the non-coding regions of our DNA are not junk.