 In case you haven't heard, you are hosting a very large and diverse population of microbes – from the top of your head to the soles of your feet and everything in between. The term used for these collections of microbes is microbiome or microbiota. This is a very hot topic of scientific research aiming to understand how the microbes around us and within us may be affecting our physiology.
In case you haven't heard, you are hosting a very large and diverse population of microbes – from the top of your head to the soles of your feet and everything in between. The term used for these collections of microbes is microbiome or microbiota. This is a very hot topic of scientific research aiming to understand how the microbes around us and within us may be affecting our physiology. We recently visited the American Museum of Natural History. I was disappointed that we forgot to purchase tickets for The Secret World Inside You exhibit. Luckily, I came across a review on CrossTalk for the new book by Susan Perkins (the curator of the exhibit) and Rob DeSalle. Welcome to the Microbiome: Getting to Know the Trillions of Bacteria and Other Microbes In, On, and Around You turned out to be a good substitute.
The book is very well written and easy to read. It is an excellent introduction to the microbiome for even the non-science type. The book covers the major topics –
starting with the basics of defining life and classifying bacteria as well as the concept of the microbiome. The book describes some of the latest research that investigates the microbes that live around us and inside us and how these collections of organisms are linked with human health and disease. Finally, they discuss what a healthy microbiome is and whether or not this is even a meaningful idea.
The microbiome research that makes headlines typically focuses on the microbiomes of everyday places and things (e.g., subways, bathrooms, kitchens) or the human microbiome and how these microbes can be linked to disease. One critical point the authors highlight is how the revolution in genome sequencing and the advent of next-generation sequencing techniques has made these types of studies possible. The scale of microbiome studies can be astounding. Researchers collect hundreds of samples and these samples can contain hundreds of different species of microbes. Using advance genome sequencing techniques, researchers can sequence and analyze these complex samples and detect even small differences in populations between the samples.
I wanted to discuss a few of the studies that the authors highlighted in their section on the microbes on and around us. My favorite was published in 2013 by Meadow et al. (coverage in outlets like Wired and The New Yorker), where the researchers examined the skin microbiomes of the players involved in a roller derby tournament. By comparing the skin microbes present on players before and after two games of this high-contact sport, they learned that the different teams started with distinct microbe populations with some variety based on the geography of the team (i.e., the team from Boston had similar microbes to the team from New York, but not San Francisco). After a jam, the teams had swapped skin microbes. The lead author on the study was a former derby skater, who thought that the sport offered a unique chance to study how microbes spread through human contact.
Another fascinating and ambitious research endeavor is the PathoMap project , which aims to categorize the microbiome in a variety of human-made environments in New York City. They started in the subway system, where they took samples from all 468 stations in NYC. They sampled ticket kiosks, turnstiles, benches, trashcans, and at places within the train itself. The first results from this study were published in Cell Systems. The results revealed an abundance of human skin bacteria as well as a rather disturbing (but frankly not surprising) number of fecal bacteria. A more recently published study examined the microbiome of the Boston subway system (original research and coverage), which showed little variation in the different subway lines even though they serve different populations. In addition, most of the microbes found were exactly what you would expect
– the bugs that typically inhabit our skin, gut, and mouths; occasionally samples from seats contained microbes usually found in the human vagina. The researchers plan to expand their study to investigate the microbial changes during cold and flu season. In addition to helping us to understand how microbes spread, these types of studies can have important implications for designing human environments to help prevent the spread of disease.
The study of microbiota is still in its infancy, so it is an exciting time to follow the research here. If you want to learn more about the microbiome, the AMNH exhibit is probably a great place to start, but if you can't get to NYC, Welcome to the Microbiome is a great alternative.
starting with the basics of defining life and classifying bacteria as well as the concept of the microbiome. The book describes some of the latest research that investigates the microbes that live around us and inside us and how these collections of organisms are linked with human health and disease. Finally, they discuss what a healthy microbiome is and whether or not this is even a meaningful idea.
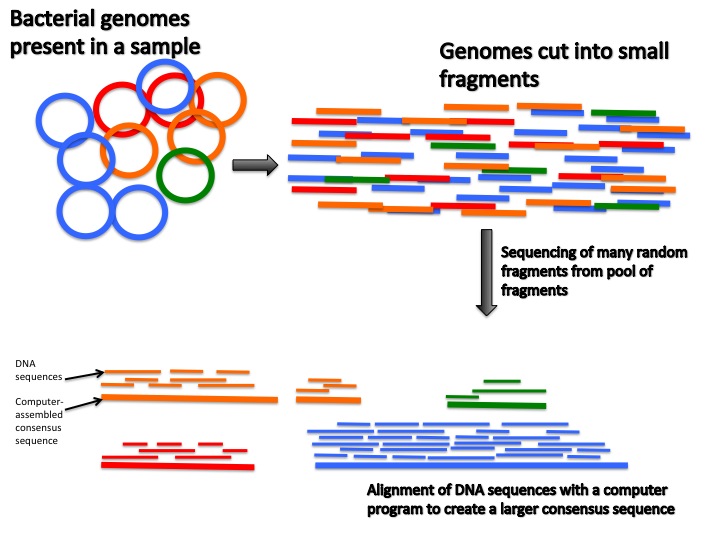 |
| Description of metagenomic sequencing from Teach the Microbiome |
I wanted to discuss a few of the studies that the authors highlighted in their section on the microbes on and around us. My favorite was published in 2013 by Meadow et al. (coverage in outlets like Wired and The New Yorker), where the researchers examined the skin microbiomes of the players involved in a roller derby tournament. By comparing the skin microbes present on players before and after two games of this high-contact sport, they learned that the different teams started with distinct microbe populations with some variety based on the geography of the team (i.e., the team from Boston had similar microbes to the team from New York, but not San Francisco). After a jam, the teams had swapped skin microbes. The lead author on the study was a former derby skater, who thought that the sport offered a unique chance to study how microbes spread through human contact.
Another fascinating and ambitious research endeavor is the PathoMap project , which aims to categorize the microbiome in a variety of human-made environments in New York City. They started in the subway system, where they took samples from all 468 stations in NYC. They sampled ticket kiosks, turnstiles, benches, trashcans, and at places within the train itself. The first results from this study were published in Cell Systems. The results revealed an abundance of human skin bacteria as well as a rather disturbing (but frankly not surprising) number of fecal bacteria. A more recently published study examined the microbiome of the Boston subway system (original research and coverage), which showed little variation in the different subway lines even though they serve different populations. In addition, most of the microbes found were exactly what you would expect
– the bugs that typically inhabit our skin, gut, and mouths; occasionally samples from seats contained microbes usually found in the human vagina. The researchers plan to expand their study to investigate the microbial changes during cold and flu season. In addition to helping us to understand how microbes spread, these types of studies can have important implications for designing human environments to help prevent the spread of disease.
 | |||||||||
| Descriptions of sampling methods in Hsu et al., 2016 ( |

